
Blogs
2024-01-22
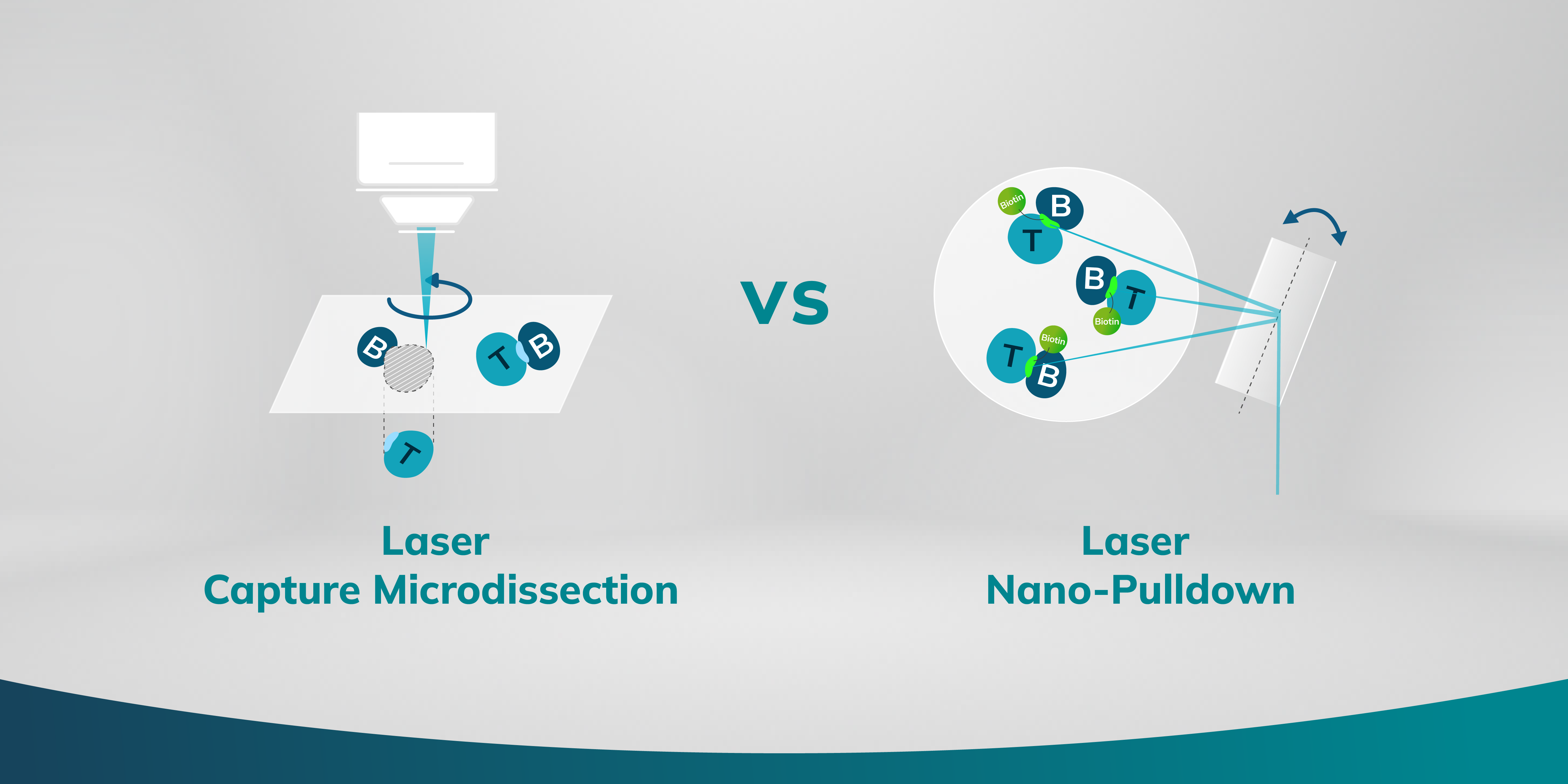
What is laser nano-pulldown? How does it differ from laser capture microdissection?
Laser Nano-Pulldown (LNP)
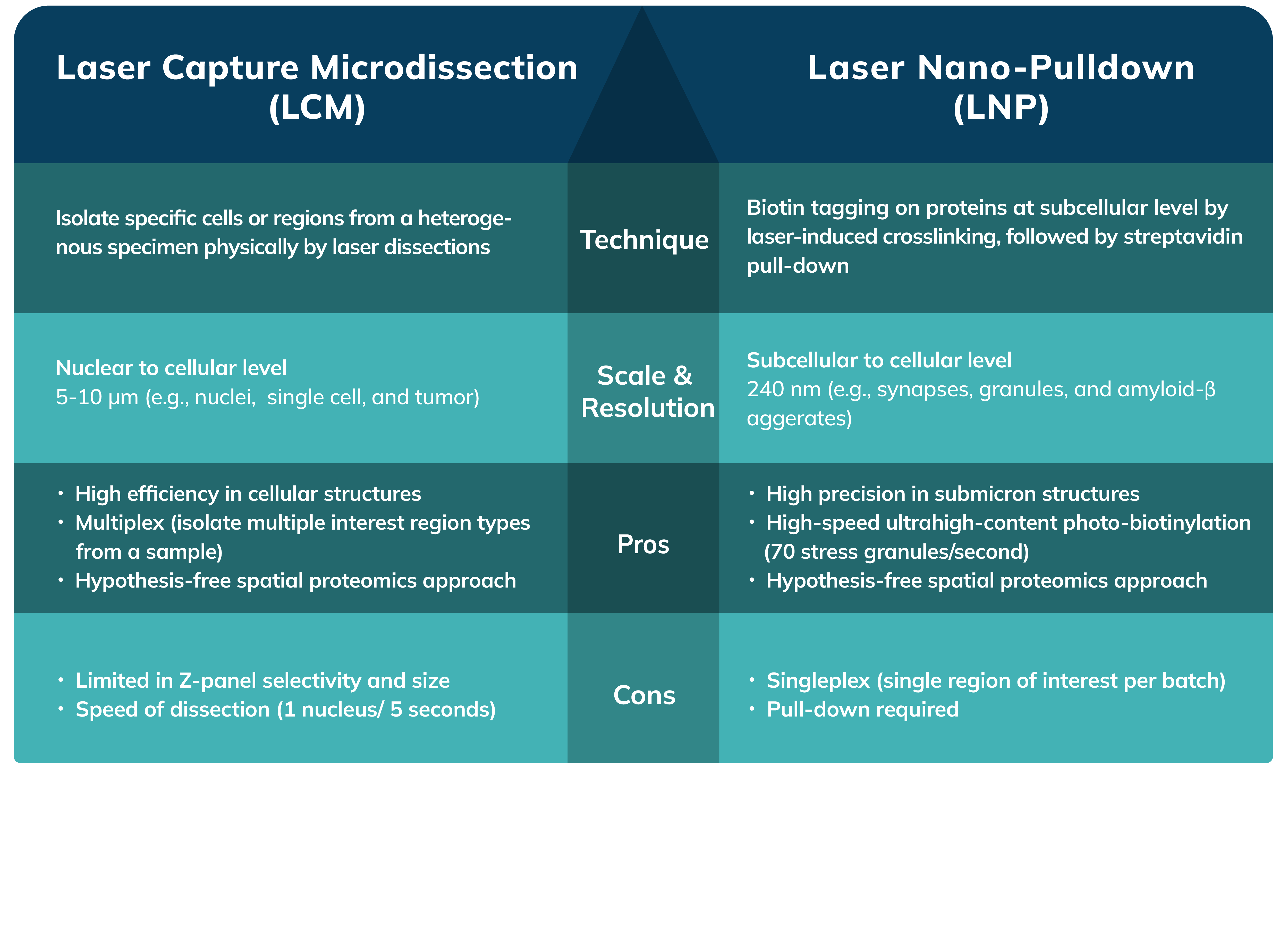
Envision a mechanism that not only facilitates the extraction of proteins under microscopic observation but does so with precision that reaches the scale of individual organelles. A new research method has surfaced, leveraging the precision of laser technology paired with an innovative nanoscale extraction process. This allows researchers to pinpoint and isolate distinct subcellular components for detailed molecular assessment. The technique, referred to as Laser Nano-Pulldown (LNP), employs an instrument called the Microscoop, renders protein isolation and identification to submicron regions within a cell.
During the LNP process, proteins situated in the immediate vicinity of the desired region of interest (ROI) undergo laser-induced biotinylation, a process that tags them with biotin. This process is automatedly repeated for thousands of fields of view to collect enough proteins for analysis, even for proteins at very low abundance. The follow-up process involves streptavidin pulldown of these biotin-tagged proteins and protein identification via liquid chromatography-mass spectrometry/mass spectrometry (LC-MS/MS), providing valuable data of the protein composition at that particular subcellular location.
Methodology
1. Targeting: A focused two-photon laser activates photochemical agents to biotinylate proteins within a precise location.
2. Binding: The activated agents attach to the proteins that are in close proximity to the target of interest.
3. Pulldown: The biotin-tagged protein are isolated through streptavidin pulldown for subsequent LC-MS/MS analysis.
Advantages and Limitations
Laser Nano-Pulldown excels at dissecting the proteome within minuscule subcellular structures. The precise control of the two-photon laser in Microscoop assure accurate axial and lateral photo-induced biotinylation, allowing target regions to be as small as 240 nm laterally and 1 um axially. In addition, with the deep penetration capability of the two-photon laser, the technique is exceptionally versatile and can be applied to a variety of cell and tissue samples. Researchers are now able to delve into small subcellular structures with unprecedented accuracy. For instance, they can isolate and examine the protein composition within the nuclei, nucleoli, and even the intricate nuclear pore complexes. The potential doesn't stop there; it extends to other structures like stress granules and primary cilia.
However, LNP does come with a constraint. Currently, it operates with one single photochemical agent, which means only one target region can be studied per sample batch, requiring additional batches for different regions of interest.
Laser Capture Microdissection (LCM)
Shifting our gaze to Laser Capture Microdissection (LCM), we encounter a technique adept at "sculpting" tissues with laser. LCM uses a laser to extract exact regions of interest from tissue samples, proving invaluable for preserving spatial context and studying complex tissues.
Methodology
1. Visualization: The sample is placed under a microscope, and the cells or areas of interest are identified visually.
2. Targeting: A specialized LCM microscope equipped with a laser is used to outline the cells or tissue regions of interest.
3. Microdissection: The laser cuts around the selected area when the laser pulses.
4. Capture: The dissected cells or tissue pieces on the transfer film are captured and are lifted out of the sample for downstream molecular analysis.
Advantages and Limitations
Laser Capture Microdissection allows precise isolation of cells from complex tissues, enabling molecular analysis for the specified spatial context. Multiple regions of interest can be extracted from a single tissue sample. For example, a researcher could separate all cell type A into one collection well, and then proceed to gather cell type B into another, enabling an organized study of different cell populations.
Nevertheless, it's important to acknowledge the limitations in spatial resolution when it comes to LCM. The smallest area that LCM can effectively isolate is approximately the size of a nucleus, or 5-10 μm. The precision in the z is dependent on the section thickness of the sample being examined. Typically, the process of excising a defined region requires about a couple of seconds. While this may not sound substantial, but when thousands of fields of view are required to collect enough low-abundant proteins, these intervals accumulate and significantly extend the duration of experiments.
Conclusion
In summary, the choice between LNP and LCM should be based on the research requirements. For investigations requiring precision at subcellular levels as small as certain organelles or protein complexes, LNP is superior. Conversely, for studies where cell populations and larger tissue regions are under scrutiny, LCM provides a more practicable solution, permitting the extraction of different cell types or regions from the same sample. The decision hinges on the specific region of interest and scale defined by the client's objectives.
It's important to note that these two techniques are not competitive but complementary, each playing to its strengths depending on the research objectives. LNP excels when subcellular molecular interactions or molecular mechanisms are of interest, whereas LCM is your go-to for wider cell populations and larger tissue sections. Together, both are de novo spatial proteomics approaches that push the boundaries of our cellular understanding and pave the way for groundbreaking discoveries in medical research and beyond.
Get In Touch
| Info@syncell.com | |
| 617-631-2746 | |
|
|
200 Dexter Ave, Watertown, MA 02472, USA |
|
|
|