
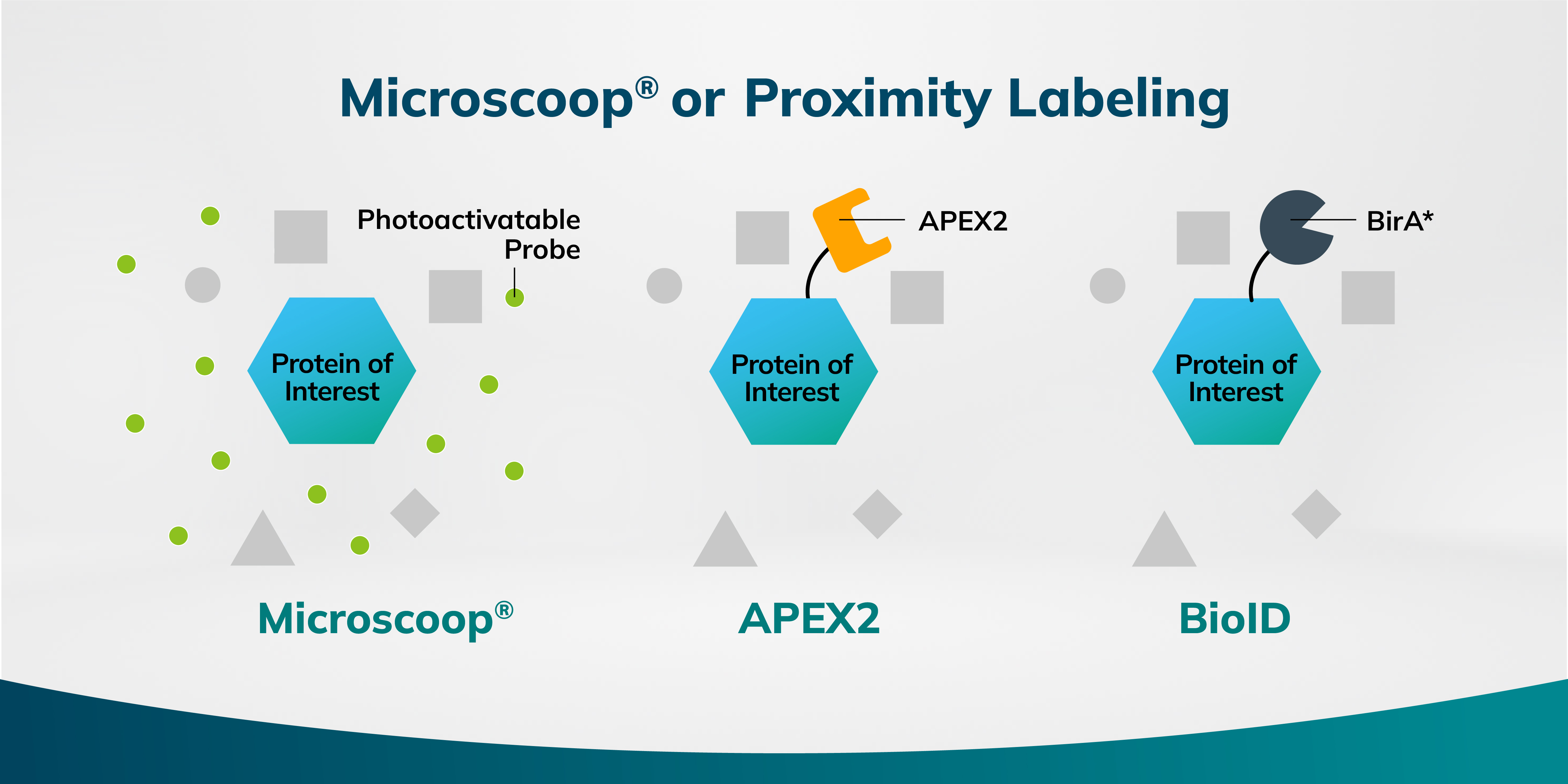
Protein-protein interactions are fundamental to cellular function, and their dysregulation underlies numerous pathological conditions. Elucidating these interactions is crucial for understanding biological processes and identifying novel therapeutic targets. To this end, several techniques are available, including Microscoop®, APEX2, and BioID. Each technique offers unique features and applications, making it suitable for specific research questions.
Understanding the Microscoop® Methodology
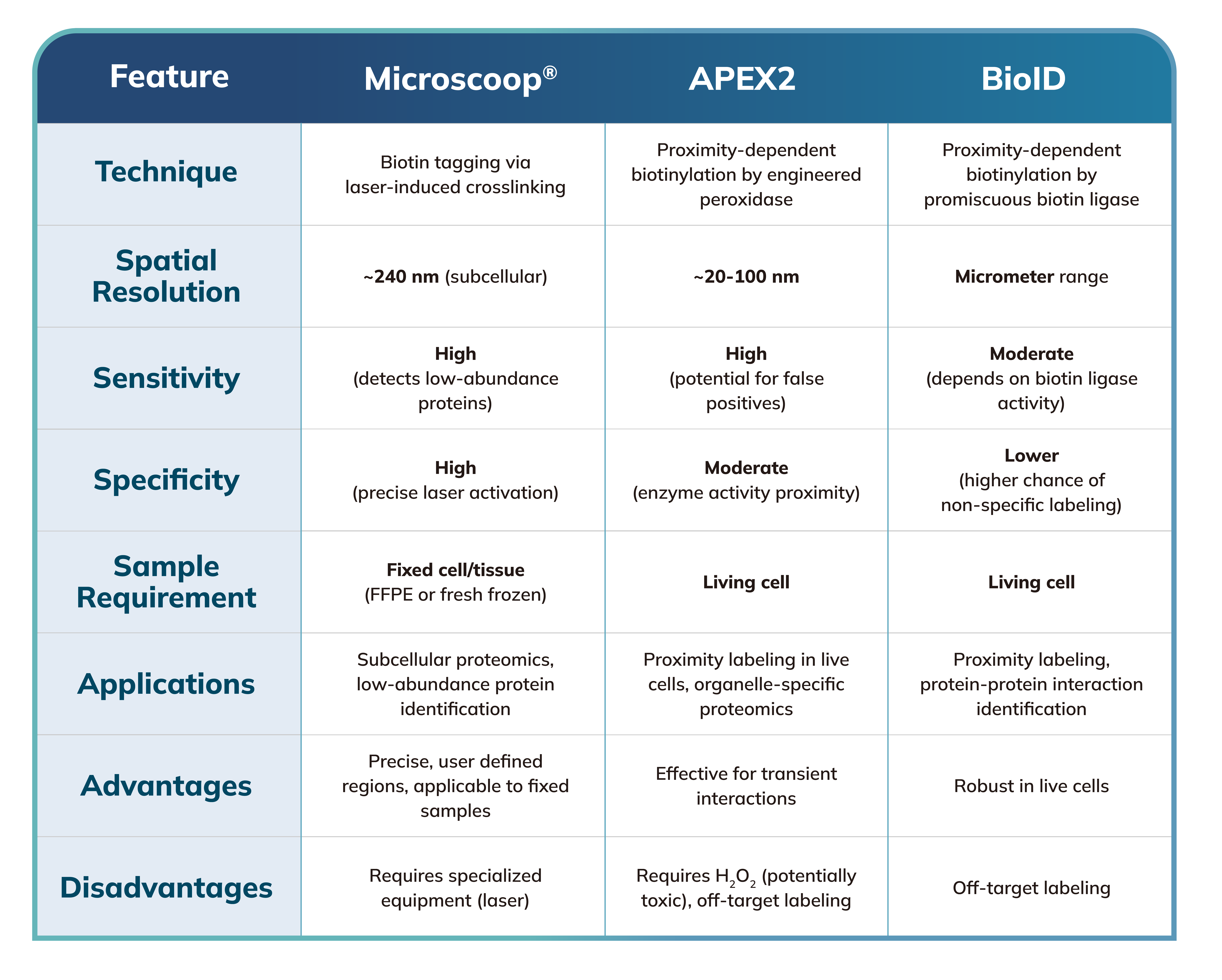
Microscoop® utilizes photo-biotinylation, a process where light induces a chemical reaction to label proteins. This method offers precise temporal and spatial control, achieving a high resolution of approximately 240 nm in protein labeling. By employing a focused two-photon laser to activate a photoactivatable biotin-based probe within user-defined regions of interest under a microscope, biotin is covalently bound to nearby proteins of the target. Microscoop® is particularly well-suited for studying fixed cells or tissues, offering advantages in terms of spatial proteomics and protein quantification. Notably, Microscoop® bypasses the need for cloning, significantly simplifying experimental workflows.
Understanding Proximity Labeling Techniques
Proximity labeling techniques such as APEX2 and BioID have emerged as powerful tools for interrogating the proteomic landscape surrounding proteins of interest.
APEX2 : A Radical-mediated Method
APEX is an engineered peroxidase designed to identify protein interacting with a protein of interest. Upon exposure to hydrogen peroxide, it catalyzes the conversion of biotin-phenol into highly reactive radicals. These radicals covalently bind to amnio acid residues on nearby proteins. This method provides a fine spatial resolution of approximately 20-100 nm but can be hindered by background noise due to the diffusion of radicals away from the target protein. While APEX2 is a valuable tool for studying protein interactions within living cells, its application is often challenged by the processes of cloning and optimization.
BioID : A biotin ligase-based method
BioID is a labeling method that utilizes a mutated form of biotin ligase, BirA*, to identify proteins in close proximity to a protein of interest. BirA* possesses a relaxed substrate specificity, allowing it to biotinylate lysine residues on nearby proteins. By fusing BirA* to a target protein, interacting proteins can be indirectly labeled. This technique is robust in live cells and less reliant on reactive intermediates. However, it has a resolution in the micrometer range and may label non-interacting proteins due to the broad range of protein labeling activity of BirA*.
Comparison of Methods
Applications : Microscoop® is versatile, applicable to both fixed cells and tissues, which are easy to handle and store. APEX2 and BioID are well-suited for investigating protein-protein interactions in living cells, while BioID is ideal for studying broad protein interaction networks.
Specificity: Microscoop® provides high specificity with reduced background noise due to precise photoactivation and limited biotinylation to the target area. APEX2 offers high resolution but may face challenges with radical diffusion, resulting in non-specific labeling of proteins. BioID's specificity can be lower owing to the promiscuous nature of BirA*.
Conclusion
While APEX2 and BioID excel at probing protein interactions within living cells, Microscoop® offers a distinct approach by employing photo-biotinylation to investigate subcellular proteomics in fixed cells and tissues. Each method possesses unique strengths, making it suitable for different research objectives based on factors such as required resolution, specificity, and sample type.

| Info@syncell.com | |
| 617-631-2746 | |
|
|
200 Dexter Ave, Watertown, MA 02472, USA |
|
|
|